The coronavirus disease 2019 (COVID-19) pandemic, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has spread with rapid devastation throughout the world. The inability to contain its spread without resorting to draconian restrictions on ordinary social interactions and travel showed the urgent need for broad-spectrum antiviral drugs to combat the virus.
Study: Efficient incorporation and template-dependent polymerase inhibition are major determinants for the broad-spectrum antiviral activity of remdesivir. Image Credit: StudioMolekuul/ Shutterstock
A new preprint suggests a fruitful approach to develop such drugs based on biochemical and molecular modeling studies of the drug remdesivir. A preprint version of the study is available on the bioRxiv* server while the article undergoes peer review.
Background
At unprecedented speed, vaccines were created using nucleic acid and adenovirus vector platforms, both of which had been the focus of research for years. These proved to be relatively safe and effective in inducing immunity against the virus. Monoclonal antibodies were also developed, showing specific neutralizing activity against SARS-CoV-2, and gained emergency use authorization.
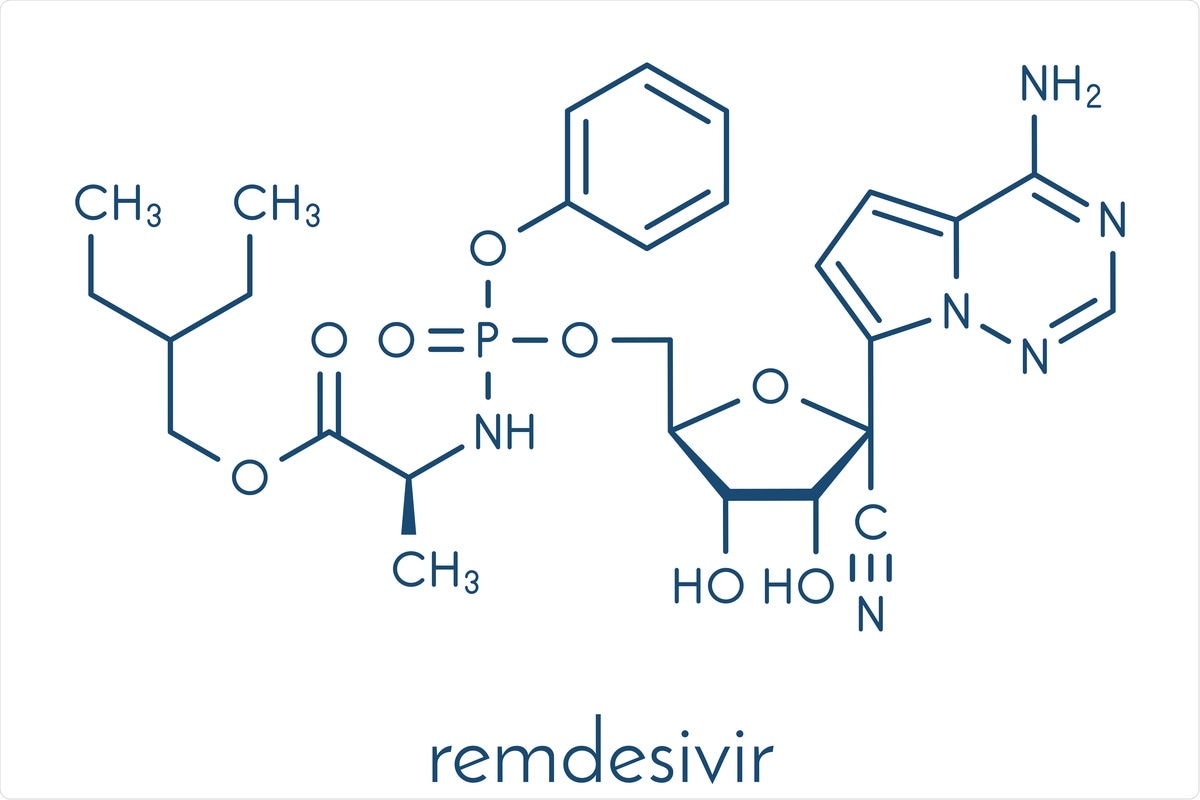
The first antiviral drug to be approved for COVID-19 treatment was the nucleoside analog prodrug remdesivir (RDV). It has shown the ability to act against several viruses in preclinical studies and has shown itself to be safe in humans during a clinical trial against the Ebola virus. Even though RDV is not very effective against Ebola, compared to several therapeutic antibodies, its safety profile led to its emergency approval for use in COVID-19 patients. However, its efficacy in this situation is very limited.
RDV showed antiviral activity against several positive-sense RNA viruses, including coronaviruses, flaviviruses like the hepatitis C virus (HCV), filoviruses like the Ebola virus, and Pneumoviruses like the respiratory syncytial virus. Others, like the Lassa virus (an arenavirus) and the flu virus (an orthomyxovirus), are not susceptible.
RDV is a modified C-adenosine monophosphate prodrug with a 1’-cyano group. In its triphosphate form (RDV-TP), it is incorporated into the RNA-dependent RNA polymerase (RdRp) of many RNA viruses, at a rate 2-3 times higher than the natural nucleoside adenosine triphosphate (ATP). After incorporation, it is present as a monophosphate (RDV-MP), and this analog at the point of incorporation “i” leads to delayed chain termination at the position “i+3”.
This can be overcome by increasing the nucleoside triphosphate (NTP). However, the embedded RDV-MP in the newly synthesized RNA becomes a template for further synthesis. This leads to reduced incorporation of the complementary uridine triphosphate (UTP) in the opposite strand, providing another inhibitory mechanism to prevent RNA synthesis and thus viral replication.
With incorporating one RDV-TP, delayed chain termination is seen only with the coronaviruses SARS-CoV, MERS-CoV, and SARS-CoV-2 RdRp complexes. A conserved serine residue clashes with the 1’-cyano group at the point i+3. With the latter, the serine-1’-cyano clash is distant from the active site, explaining why NTP concentrations > 10 μM were often successful at overcoming the inhibition.
For SARS-CoV-2 RdRp, the concentration of nucleotides had to be ten-fold higher to overcome the inhibitory effect by 50% beyond point i, relative to HCV RdRp.
Earlier, researchers have shown that when RDV-TP is incorporated at multiple consecutive positions, delayed chain-termination at positions “i+3” and “i+5” occurs even with Ebola, RSV, and Nipah virus RdRp complexes.
What did the study show?
In the current study, the researchers found that the antiviral activity of this drug was a function of two actions.
Firstly, they found that the drug’s ability to inhibit the virus in a cell-based assay was closely linked to the rate of incorporation of RDV-TP.
The efficiency of selective incorporation of RDV-TP is measured by the ratio of efficiency of incorporation of the original ATP in preference to the analogous RDV-TP. For SARS-CoV-2, the ratio is 0.3, comparable to HCV (0.9), Ebola virus (4.0), and RSV enzymes (2.7). In contrast, the Lassa virus had a ratio of 20, and the flu virus, as did other viruses that were not inhibited by the drug, indicating that these viruses did not incorporate RDV-TP efficiently.
This difference in incorporation by viral RdRp depends to a large extent on differences in the residues that make up the nucleotide-binding site, which is highly conserved and includes several motifs. Of these, motifs A, B, and C are key in their interactions with the newly incorporated NTP.
The authors then created a model of the ternary structure of the binding sites of the RdRp enzymes from a panel of viruses. This reflected the observed difference in binding of RDV-TP to SARS-CoV-2 and HCV RdRp enzymes, compared to the Lassa virus or the flu virus RdRp.
The former pair had very similar binding sites for RDV-TP, requiring almost no change in position or recognition of the ribose sugar molecule compared to ATP binding. The 1’-cyano group can bind easily to the polar 1′ binding pocket.
With the SARS-CoV-2 RdRp, the incorporation of RDV-TP caused delayed chain termination at position i+3. With the flu virus, the portion of the active site containing polar residues is too far from that which recognizes the ribose to offer a similarly favorable interaction with the NTP. A water molecule also appears to be present at the binding site for the nucleotide. However, in the presence of the 1’-cyano group, the water would be displaced.
The active sites of the Lassa and flu viruses appear to have similar sequences, explaining the low rate of RDV-TP incorporation by these RdRp enzymes.
Secondly, in HCV RdRp, this led to a slight reduction in RNA synthesis at the point of incorporation of the altered nucleoside (i) but no inhibition at a point 3 nucleotides ahead (i+3). Moreover, when the succeeding nucleoside triphosphates (NTP) were low in concentration, the inhibitory effect at the point of incorporation was rapidly overcome, and RNA synthesis proceeded.
Thus, delayed chain termination was not significant in other viral polymerases, indicating that RDV does not suppress primer extension reactions to any great extent.
The question remained as to the mechanism by which RDV could exert inhibitory effects on a diverse range of viruses. Even though both HCV and SARS-CoV-2 showed the ability to incorporate RDV-TP into the RNA strand very efficiently, only with the latter was RdRp inhibited to any significant extent, as seen by the blocking of primer extension.
In other words, the delayed chain termination may contribute to the antiviral effect of RDV-TP in coronaviruses. Still, another mechanism must be sought for the observed inhibition of HCV as well.
This is explained by the inhibition of both SARS-CoV-2 and HCV RdRp before the point where UTP is incorporated, which is overcome by increasing the UTP concentration. However, the inhibitory effect is more easily overcome with HCV RdRp, which also shows inhibition only at position 10, whereas SARS-CoV-2 RdRp is inhibited at position ten and its neighboring positions.
Both these mechanisms combine to enhance inhibition of RNA synthesis for SARS-CoV-2 more than for HCV. While primer extension inhibition is weak, variable, and easily overcome by higher NTP concentrations, the template-dependent inhibition of UTP incorporation opposite the embedded RDV-MP is prominent for both viruses.
What are the implications?
In clinical studies, RDV has not shown the ability to reduce mortality due to COVID-19, but the recovery time has been shortened. The second disadvantage of RDV is the need for intravenous administration. Oral drugs could be administered on an outpatient basis, thus conceivably reducing the incidence of severe COVID-19 by inhibiting disease progression.
The findings of studies such as this one could help understand the molecular basis of antiviral activity and thus accelerate the development of oral antivirals with a broad spectrum of activity.
Overall, this analysis suggests that the inhibitory effect in primer extension reactions is heterogeneous and generally weak. In contrast, the template-dependent inhibition of RNA synthesis seems to provide a unifying mechanism that shows strong inhibition of UTP incorporation opposite RDV-MP.”
Combined with earlier data, the researchers conclude that the antiviral effect of RDV is correlated with the efficient incorporation of RDV-TP into the newly formed RNA, and this selective incorporation is essential for its inhibitory activity. The type of RdRp determines the susceptibility of the RNA to chain termination, but overall, the inhibition by this mechanism is relatively easily overcome by increasing the NTP concentrations.
This is because the higher concentrations allow the enzyme to translocate and continue RNA synthesis. The authors suggest that this type of weak inhibition of the synthesis of the first RNA strand could be desirable for the coronavirus as it allows for immune evasion.
However, the template-dependent inhibition of RNA synthesis opposite the embedded RDV-MP is seen with the panel of RdRp enzymes studied here. The inhibition of UTP incorporation opposite the RDV-MP is probably the result of steric hindrances caused by the presence of the 1’-cyano group, which clashes with the conserved motif F residues.
This mechanism of inhibition is applicable across a wide spectrum of viruses, even those that are not susceptible to this drug. However, it is not sufficient to cause antiviral efficacy, which depends on the efficient selective incorporation of RDV-TP.
There is no significant inhibition by RDV-TP in primer extensions, and strong inhibition or even termination when RDV-MP is embedded in the template. However, inhibition translates to antiviral effects only if the rate of incorporation of RDV-TP is sufficiently high.”
This offers the opportunity to develop a drug that can be incorporated efficiently by viral RdRp enzymes while exploiting the steric effects of a bulky group at the 1’-position.
*Important notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information
- Gordon, C. J. et al. (2021). Efficient Incorporation and Template-Dependent Polymerase Inhibition Are Major Determinants for The Broad-Spectrum Antiviral Activity of Remdesivir. bioRxiv preprint. doi: https://doi.org/10.1101/2021.10.14.464416.